The OG Clever Machine Has Moved
Hello! The Clever Machine has moved! All new material will be posted to the new site, hosted on GitHub pages, where I will continue to provide similar content on analysis techniques, algorithms, theory, or things I think are cool.
Also, because the data science, machine learning, and analytics fields are now so heavily based in open-source software like Python, I will migrating lot of the original material from this WordPress site to the new site, but translating the MATLAB implementations into Python 3.
While in the process of migrating content to the new site, I’ll leave this site up, but once I’ve migrated all posts, The OG Clever Machine will shut down. If you like, you can still access old archives of the OG Clever Machine via the Wayback Machine.
Looking forward to seeing you all on the new blog site!
Derivation: Maximum Likelihood for Boltzmann Machines
In this post I will review the gradient descent algorithm that is commonly used to train the general class of models known as Boltzmann machines. Though the primary goal of the post is to supplement another post on restricted Boltzmann machines, I hope that those readers who are curious about how Boltzmann machines are trained, but have found it difficult to track down a complete or straight-forward derivation of the maximum likelihood learning algorithm for these models (as I have), will also find the post informative.
First, a little background: Boltzmann machines are stochastic neural networks that can be thought of as the probabilistic extension of the Hopfield network. The goal of the Boltzmann machine is to model a set of observed data in terms of a set of visible random variables and a set of latent/unobserved random variables
. Due to the relationship between Boltzmann machines and neural networks, the random variables are often are often referred to as “units.” The role of the visible units is to approximate the true distribution of the data, while the role of the latent variables it to extend the expressiveness of the model by capturing underlying features in the observed data. The latent variables are often referred to as hidden units, as they do not result directly from the observed data and are generally marginalized over to obtain the likelihood of the observed data, i.e.
,
where is the joint probability distribution over the visible and hidden units based on the current model parameters
. The general Boltzmann machine defines
through a set of weighted, symmetric connections between all visible and hidden units (but no connections from any unit to itself). The graphical model for the general Boltzmann machine is shown in Figure 1.
Given the current state of the visible and hidden units, the overall configuration of the model network is described by a connectivity function , parameterized by
:
The parameter matrix defines the connection strength between the visible and hidden units. The parameters
and
define the connection strength amongst hidden units and visible units, respectively. The model also includes a set of biases
and
that capture offsets for each of the hidden and visible units.
The Boltzmann machine has been used for years in field of statistical mechanics to model physical systems based on the principle of energy minimization. In the statistical mechanics, the connectivity function is often referred to the “energy function,” a term that is has also been standardized in the statistical learning literature. Note that the energy function returns a single scalar value for any configuration of the network parameters and random variable states.
Given the energy function, the Boltzmann machine models the joint probability of the visible and hidden unit states as a Boltzmann distribution:
The partition function is a normalizing constant that is calculated by summing over all possible states of the network
. Here we assume that all random variables take on discrete values, but the analogous derivation holds for continuous or mixed variable types by replacing the sums with integrals accordingly.
The common way to train the Boltzmann machine is to determine the parameters that maximize the likelihood of the observed data. To determine the parameters, we perform gradient descent on the log of the likelihood function (In order to simplify the notation in the remainder of the derivation, we do not include the explicit dependency on the parameters . To further simplify things, let’s also assume that we calculate the gradient of the likelihood based on a single observation.):
The gradient calculation is as follows:
Here we can simplify the expression somewhat by noting that , that
, and also that
is a constant:
If we also note that , and use the definition of conditional probability
, we can further simplify the expression for the gradient:
Here is the expected value under the distribution
. Thus the gradient of the likelihood function is composed of two parts. The first part is expected gradient of the energy function with respect to the conditional distribution
. The second part is expected gradient of the energy function with respect to the joint distribution over all variable states. However, calculating these expectations is generally infeasible for any realistically-sized model, as it involves summing over a huge number of possible states/configurations. The general approach for solving this problem is to use Markov Chain Monte Carlo (MCMC) to approximate these sums:
Here is the sample average of samples drawn according to the process
. The first term is calculated by taking the average value of the energy function gradient when the visible and hidden units are being driven by observed data samples. In practice, this first term is generally straightforward to calculate. Calculating the second term is generally more complicated and involves running a set of Markov chains until they reach the current model’s equilibrium distribution (i.e. via Gibbs sampling, Metropolis-Hastings, or the like), then taking the average energy function gradient based on those samples. See this post on MCMC methods for details. It turns out that there is a subclass of Boltzmann machines that, due to a restricted connectivity/energy function (specifically, the parameters
), allow for efficient MCMC by way of blocked Gibbs sampling. These models, known as restricted Boltzman machines have become an important component for unsupervised pretraining in the field of deep learning and will be the focus of a related post.
A Gentle Introduction to Artificial Neural Networks
The material in this post has been migrated to a post by the same name on my github pages website.
Derivation: Derivatives for Common Neural Network Activation Functions
The material in this post has been migraged with python implementations to my github pages website.
Derivation: Error Backpropagation & Gradient Descent for Neural Networks
The material in this post has been migraged with python implementations to my github pages website.
Model Selection: Underfitting, Overfitting, and the Bias-Variance Tradeoff
The material in this post has been migrated with python implementations to my github pages website.
The Statistical Whitening Transform
In a number of modeling scenarios, it is beneficial to transform the to-be-modeled data such that it has an identity covariance matrix, a procedure known as Statistical Whitening. When data have an identity covariance, all dimensions are statistically independent, and the variance of the data along each of the dimensions is equal to one. (To get a better idea of what an identity covariance entails, see the following post.)
Enforcing statistical independence is useful for a number of reasons. For example, in probabilistic models of data that exist in multiple dimensions, the joint distribution–which may be very complex and difficult to characterize–can factorize into a product of many simpler distributions when the dimensions are statistically independent. Forcing all dimensions to have unit variance is also useful. For instance, scaling all variables to have the same variance treats each dimension with equal importance.
In the remainder of this post we derive how to transform data such that it has an identity covariance matrix, give some examples of applying such a transformation to real data, and address some interpretations of statistical whitening in the scope of theoretical neuroscience.
Decorrelation: Transforming Data to Have a Diagonal Covariance Matrix
Let’s say we have some data matrix composed of
dimensions and
observations (
has size
). Let’s also assume that the rows of
have been centered (the mean has been subracted across all observations) . The covariance
of each of the dimensions with respect to the other is
(1)
Where the covariance can be estimated from the data matrix as follows:
(2)
The covariance matrix , by definition (Equation 2) is symmetric and positive semi-definite (if you don’t know what that means, don’t worry it’s not terribly important for this discussion). Thus we can write the matrix as the product of two simpler matrices
and
, using a procedure known as Eigenvalue Decomposition:
(3)
The matrix is an
-sized matrix, where each column is an eigenvector of
, and
is a diagonal matrix whose diagonal elements
are eigenvalues that correspond to the eigenvectors of the
-th column of
. For more details on eigenvectors and eigenvalues see the following. From Equation (3), and using a little algebra, we can transform
into the diagonal matrix
(4)
Now, imagine the goal is to transform the data matrix into a new data matrix
(5)
whose dimensions are uncorrelated (i.e. has a diagonal covariance
). Thus we want to determine the transformation
that makes:
(6)
Here we derive the expression for using Equations (2), (4), (5), and (6):
(a la Equations (5) and (6))
(via Equation (2))
(via Equation (4))
now, because (see following link for details)
and thus
(7)
This means that we can transform into an uncorrelated (i.e. orthogonal) set of variables by premultiplying data matrix
with the transpose of the the eigenvectors of data covariance matrix
.
Whitening: Transforming data to have an Identity Covariance matrix
Ok, so now we have a way of transforming our data so that the dimensions are uncorrelated. However, this only gives us a diagonal covariance matrix, not an Identity covariance matrix. In order to obtain an Identity covariance, we also need to scale each dimension so that its variance is equal to one. How can we determine this transformation? We know how to transform our data so that the covariance is equal to . If we can determine the transformation that leaves
, then we can apply this transformation to our decorrelated covariance to give us the desired whitening transform. We can determine this from the somewhat trivial notion that
(8)
and further that
(9)
Now, using Equation (4) along with Equation (8), we can see that
(10)
Now say that we define a variable , where
is the desired whitening transform, that leaves the covariance of
equal to the identity matrix. Using essentially the same set of derivation steps as above to solve for
, but starting from Equation (9) we find that
(11)
(12)
Thus, the whitening transform is simply the decorrelation transform, but scaled by the inverse of the square root of the (here the inverse and square root can be performed element-wise because
is a diagonal matrix).
Interpretation of the Whitening Transform
So what does the whitening transformation actually do to the data (below, blue points)? We investigate this transformation below: The first operation decorrelates the data by premultiplying the data with the eigenvector matrix , calculated from the data covariance. This decorrelation can be thought of as a rotation that reorients the data so that the principal axes of the data are aligned with the axes along which the data has the largest (orthogonal) variance. This rotation is essentially the same procedure as the oft-used Principal Components Analysis (PCA), and is shown in the middle row.
The second operation, scaling by can be thought of squeezing the data–if the variance along a dimension is larger than one–or stretching the data–if the variance along a dimension is less than one. The stretching and squeezing forms the data into a sphere about the origin (which is why whitening is also referred to as “sphering”). This scaling operation is depicted in the bottom row in the plot above.
The MATLAB to make make the plot above is here:
% INITIALIZE SOME CONSTANTS
mu = [0 0];
S = [1 .9; .9 3];
% SAMPLE SOME DATAPOINTS
nSamples = 1000;
samples = mvnrnd(mu,S,nSamples)';
% WHITEN THE DATA POINTS...
[E,D] = eig(S);
% ROTATE THE DATA
samplesRotated = E*samples;
% TAKE D^(-1/2)
D = diag(diag(D).^-.5);
% SCALE DATA BY D
samplesRotatedScaled = D*samplesRotated;
% DISPLAY
figure;
subplot(311);
plot(samples(1,:),samples(2,:),'b.')
axis square, grid
xlim([-5 5]);ylim([-5 5]);
title('Original Data');
subplot(312);
plot(samplesRotated(1,:),samplesRotated(2,:),'r.'),
axis square, grid
xlim([-5 5]);ylim([-5 5]);
title('Decorrelate: Rotate by V');
subplot(313);
plot(samplesRotatedScaled(1,:),samplesRotatedScaled(2,:),'ko')
axis square, grid
xlim([-5 5]);ylim([-5 5]);
title('Whiten: scale by D^{-1/2}');
The transformation in Equation (11) and implemented above whitens the data but leaves the data aligned with principle axes of the original data. In order to observe the data in the original space, it is often customary “un-rotate” the data back into it’s original space. This is done by just multiplying the whitening transform by the inverse of the rotation operation defined by the eigenvector matrix. This gives the whitening transform:
(13)
Let’s take a look an example of using statistical whitening for a more complex problem: whitening patches of images sampled from natural scenes.
Example: Whitening Natural Scene Image Patches
Modeling the local spatial structure of pixels in natural scene images is important in many fields including computer vision and computational neuroscience. An interesting model of natural scenes is one that can account for interesting, high-order statistical dependencies between pixels. However, because natural scenes are generally composed of continuous objects or surfaces, a vast majority of the spatial correlations in natural image data can be explained by local pairwise dependencies. For example, observe the image below.
% LOAD AND DISPLAY A NATURAL IMAGE
im = double(imread('cameraman.tif'));
figure
imagesc(im); colormap gray; axis image; axis off;
title('Base Image')
Given one of the gray pixels in the upper portion of the image, it is very likely that all pixels within the local neighborhood will also be gray. Thus there is a large amount of correlation between pixels in local regions of natural scenes. Statistical models of local structure applied to natural scenes will be dominated by these pairwise correlations, unless they are removed by preprocessing. Whitening provides such a preprocessing procedure.
Below we create and display a dataset of local image patches of size extracted at random from the image above. Each patch is rastered out into a column vector of size
. Each of these patches can be thought of as samples of the local structure of this natural scene. Below we use the whitening transformation to remove pairwise correlations between pixels in each patch and scale the variance of each pixel to be one.
On the left is the dataset of extracted image patches, along with the corresponding covariance matrix for the image patches on the right. The large local correlation within the neighborhood of each pixel is indicated by the large bright diagonal regions throughout the covariance matrix.
The MATLAB code to extract and display the patches shown above is here:
% CREATE PATCHES DATASET FROM NATURAL IMAGE
rng(12345)
imSize = 256;
nPatches = 400; % (MAKE SURE SQUARE)
patchSize = 16;
patches = zeros(patchSize*patchSize,nPatches);
patchIm = zeros(sqrt(nPatches)*patchSize);
% PAD IMAGE FOR EDGE EFFECTS
im = padarray(im,[patchSize,patchSize],'symmetric');
% EXTRACT PATCHES...
for iP = 1:nPatches
pix = ceil(rand(2,1)*imSize);
rows = pix(1):pix(1)+patchSize-1;
cols = pix(2):pix(2)+patchSize-1;
tmp = im(rows,cols);
patches(:,iP) = reshape(tmp,patchSize*patchSize,1);
rowIdx = (ceil(iP/sqrt(nPatches)) - 1)*patchSize + ...
1:ceil(iP/sqrt(nPatches))*patchSize;
colIdx = (mod(iP-1,sqrt(nPatches)))*patchSize+1:patchSize* ...
((mod(iP-1,sqrt(nPatches)))+1);
patchIm(rowIdx,colIdx) = tmp;
end
% CENTER IMAGE PATCHES
patchesCentered = bsxfun(@minus,patches,mean(patches,2));
% CALCULATE COVARIANCE MATRIX
S = patchesCentered*patchesCentered'/nPatches;
% DISPLAY PATCHES
figure;
subplot(121);
imagesc(patchIm);
axis image; axis off; colormap gray;
title('Extracted Patches')
% DISPLAY COVARIANCE
subplot(122);
imagesc(S);
axis image; axis off; colormap gray;
title('Extracted Patches Covariance')
Below we implement the whitening transformation described above to the extracted image patches and display the whitened patches that result.
 On the left, we see that the whitening procedure zeros out all areas in the extracted patches that have the same value (zero is indicated by gray). The whitening procedure also boosts the areas of high-contrast (i.e. edges). The right plots the covariance matrix for the whitened patches. The covarance matrix is diagonal, indicating that pixels are now independent. In addition, all diagonal entries have the same value, indicating the that all pixels now have the same variance (i.e. 1). The MATLAB code used to whiten the image patches and create the display above is here:
On the left, we see that the whitening procedure zeros out all areas in the extracted patches that have the same value (zero is indicated by gray). The whitening procedure also boosts the areas of high-contrast (i.e. edges). The right plots the covariance matrix for the whitened patches. The covarance matrix is diagonal, indicating that pixels are now independent. In addition, all diagonal entries have the same value, indicating the that all pixels now have the same variance (i.e. 1). The MATLAB code used to whiten the image patches and create the display above is here:
%% MAIN WHITENING
% DETERMINE EIGENECTORS & EIGENVALUES
% OF COVARIANCE MATRIX
[E,D] = eig(S);
% CALCULATE D^(-1/2)
d = diag(D);
d = real(d.^-.5);
D = diag(d);
% CALCULATE WHITENING TRANSFORM
W = E*D*E';
% WHITEN THE PATCHES
patchesWhitened = W*patchesCentered;
% DISPLAY THE WHITENED PATCHES
wPatchIm = zeros(size(patchIm));
for iP = 1:nPatches
rowIdx = (ceil(iP/sqrt(nPatches)) - 1)*patchSize + 1:ceil(iP/sqrt(nPatches))*patchSize;
colIdx = (mod(iP-1,sqrt(nPatches)))*patchSize+1:patchSize* ...
((mod(iP-1,sqrt(nPatches)))+1);
wPatchIm(rowIdx,colIdx) = reshape(patchesWhitened(:,iP),...
[patchSize,patchSize]);
end
figure
subplot(121);
imagesc(wPatchIm);
axis image; axis off; colormap gray; caxis([-5 5]);
title('Whitened Patches')
subplot(122);
imagesc(cov(patchesWhitened'));
axis image; axis off; colormap gray; %colorbar
title('Whitened Patches Covariance');
Investigating the Whitening Matrix: implications for theoretical neuroscience
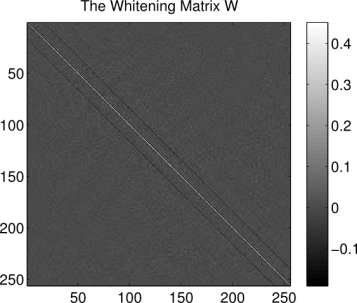
So what does the whitening matrix look like, and what does it do? Below is the whitening matrix calculated for the image patches dataset:
% DISPLAY THE WHITENING MATRIX
figure; imagesc(W);
axis image; colormap gray; colorbar
title('The Whitening Matrix W')
Each column of is the operation that scales the variance of the corresponding pixel to be equal to one and forces that pixel independent of the others in the
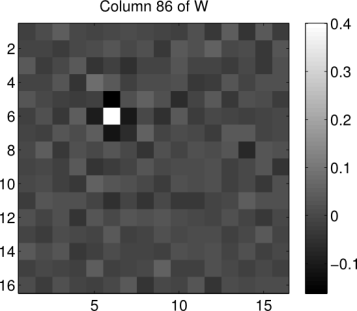
patch. So what exactly does such an operation look like? We can get an idea by reshaping a column of W back into the shape of the image patches. Below we show what the 86th column of W looks like when reshaped in such a way (the index 86 has no particular significance, it was chosen at random):
% DISPLAY A COLUMN OF THE WHITENING MATRIX
figure; imagesc(reshape(W(:,86),16,16)),
colormap gray,
axis image, colorbar
title('Column 86 of W')
We see that the operation is essentially an impulse centered on the 86th pixel in the image (counting pixels starting in the upper left corner, proceeding down columns). This impulse is surrounded by inhibitory weights. If we were to look at the remaining columns of , we would find that that the same center-surround operation is being replicated at every pixel location in each image patch. Essentially, the whitening transformation is performing a convolution of each image patch with a center-surround filter whose properties are estimated from the patches dataset. Similar techniques are common in computer vision edge-detection algorithms.
Implications for theoretical neuroscience
A theoretical function of the primate retina is data compression: a large number of photoreceptors pass data from the retina into a physiological bottleneck, the optic nerve, which has far fewer fibers than retinal photoreceptors. Thus removing redundant information is an important task that the retina must perform. When observing the whitened image patches above, we see that redundant information is nullified; pixels that have similar local values to one another are zeroed out. Thus, statistical whitening is a viable form of data compression
It turns out that there is a large class of ganglion cells in the retina whose spatial receptive fields exhibit…that’s right center-surround activation-inhibition like the operation of the whitening matrix shown above! Thus it appears that the primate visual system may be performing data compression at the retina by means of a similar operation to statistical whitening. Above, we derived the center-surround whitening operation based on data sampled from a natural scene. Thus it is seems reasonable that the primate visual system may have evolved a similar data-compression mechanism through experience with natural scenes, either through evolution, or development.
Covariance Matrices and Data Distributions
Correlation between variables in a -dimensional dataset are often summarized by a
covariance matrix. To get a better understanding of how correlation matrices characterize correlations between data points, we plot data points drawn from 3 different 2-dimensional Gaussian distributions, each of which is defined by a different covariance matrix.
The left plots below display the covariance matrix for each Gaussian distribution. The values along the diagonal represent the variance of the data along each dimension, and the off-diagonal values represent the covariances between the dimensions. Thus the
-th entry of each matrix represents the correlation between the
-th and
-th dimensions. The right plots show data drawn from the corresponding 2D Gaussian.
The top row plot display a covariance matrix equal to the identity matrix, and the points drawn from the corresponding Gaussian distribution. The diagonal values are 1, indicating the data have variance of 1 along both of the dimensions. Additionally, the off-diagonal elements are zero, meaning that the two dimensions are uncorrelated. We can see this in the data drawn from the distribution as well. The data are distributed in a sphere about origin. For such a distribution of points, it is difficult (impossible) to draw any single regression line that can predict the second dimension from the first, and vice versa. Thus an identity covariance matrix is equivalent to having independent dimensions, each of which has unit (i.e. 1) variance. Such a dataset is often called “white” (this naming convention comes from the notion that white noise signals–which can be sampled from independent Gaussian distributions–have equal power at all frequencies in the Fourier domain).
The middle row plots the points that result from a diagonal, but not identity covariance matrix. The off-diagonal elements are still zero, indicating that the dimensions are uncorrelated. However, the variances along each dimension are not equal to one, and are not equal. This is demonstrated by the elongated distribution in red. The elongation is along the second dimension, as indicated by the larger value in the bottom-right (point ) of the covariance matrix.
The bottom row plots points that result from a non-diagonal covariance matrix. Here the off-diagonal elements of covariance matrix have non-zero values, indicating a correlation between the dimensions. This correlation is reflected in the distribution of drawn datapoints (in blue). We can see that the primary axis along which the points are distributed is not along either of the dimensions, but a linear combination of the dimensions.
The MATLAB code to create the above plots is here
% INITIALIZE SOME CONSTANTS
mu = [0 0]; % ZERO MEAN
S = [1 .9; .9 3]; % NON-DIAGONAL COV.
SDiag = [1 0; 0 3]; % DIAGONAL COV.
SId = eye(2); % IDENTITY COV.
% SAMPLE SOME DATAPOINTS
nSamples = 1000;
samples = mvnrnd(mu,S,nSamples)';
samplesId = mvnrnd(mu,SId,nSamples)';
samplesDiag = mvnrnd(mu,SDiag,nSamples)';
% DISPLAY
subplot(321);
imagesc(SId); axis image,
caxis([0 1]), colormap hot, colorbar
title('Identity Covariance')
subplot(322)
plot(samplesId(1,:),samplesId(2,:),'ko'); axis square
xlim([-5 5]), ylim([-5 5])
grid
title('White Data')
subplot(323);
imagesc(SDiag); axis image,
caxis([0 3]), colormap hot, colorbar
title('Diagonal Covariance')
subplot(324)
plot(samplesDiag(1,:),samplesDiag(2,:),'r.'); axis square
xlim([-5 5]), ylim([-5 5])
grid
title('Uncorrelated Data')
subplot(325);
imagesc(S); axis image,
caxis([0 3]), colormap hot, colorbar
title('Non-diagonal Covariance')
subplot(326)
plot(samples(1,:),samples(2,:),'b.'); axis square
xlim([-5 5]), ylim([-5 5])
grid
title('Correlated Data')
fMRI In Neuroscience: Efficiency of Event-related Experiment Designs
Event-related fMRI experiments are used to detect selectivity in the brain to stimuli presented over short durations. An event is generally modeled as an impulse function that occurs at the onset of the stimulus in question. Event-related designs are flexible in that many different classes of stimuli can be intermixed. These designs can minimize confounding behavioral effects due to subject adaptation or expectation. Furthermore, stimulus onsets can be modeled at frequencies that are shorter than the repetition time (TR) of the scanner. However, given such flexibility in design and modeling, how does one determine the schedule for presenting a series of stimuli? Do we space out stimulus onsets periodically across a scan period? Or do we randomize stimulus onsets? Furthermore what is the logic for or against either approach? Which approach is more efficient for gaining incite into the selectivity in the brain?
Simulating Two fMRI Experiments: Periodic and Random Stimulus Onsets
To get a better understanding of the problem of choosing efficient experiment design, let’s simulate two simple fMRI experiments. In the first experiment, a stimulus is presented periodically 20 times, once every 4 seconds, for a run of 80 seconds in duration. We then simulate a noiseless BOLD signal evoked in a voxel with a known HRF. In the second experiment, we simulate the noiseless BOLD signal evoked by 20 stimulus onsets that occur at random times over the course of the 80 second run duration. The code for simulating the signals and displaying output are shown below:
rand('seed',12345);
randn('seed',12345);
TR = 1 % REPETITION TIME
t = 1:TR:20; % MEASUREMENTS
h = gampdf(t,6) + -.5*gampdf(t,10); % ACTUAL HRF
h = h/max(h); % SCALE TO MAX OF 1
% SOME CONSTANTS...
trPerStim = 4; % # TR PER STIMULUS FOR PERIODIC EXERIMENT
nRepeat = 20; % # OF TOTAL STIMULI SHOWN
nTRs = trPerStim*nRepeat
stimulusTrain0 = zeros(1,nTRs);
beta = 3; % SELECTIVITY/HRF GAIN
% SET UP TWO DIFFERENT STIMULUS PARADIGM...
% A. PERIODIC, NON-RANDOM STIMULUS ONSET TIMES
D_periodic = stimulusTrain0;
D_periodic(1:trPerStim:trPerStim*nRepeat) = 1;
% UNDERLYING MODEL FOR (A)
X_periodic = conv2(D_periodic,h);
X_periodic = X_periodic(1:nTRs);
y_periodic = X_periodic*beta;
% B. RANDOM, UNIFORMLY-DISTRIBUTED STIMULUS ONSET TIMES
D_random = stimulusTrain0;
randIdx = randperm(numel(stimulusTrain0)-5);
D_random(randIdx(1:nRepeat)) = 1;
% UNDERLYING MODEL FOR (B)
X_random = conv2(D_random,h);
X_random = X_random(1:nTRs);
y_random = X_random*beta;
% DISPLAY STIMULUS ONSETS AND EVOKED RESPONSES
% FOR EACH EXPERIMENT
figure
subplot(121)
stem(D_periodic,'k');
hold on;
plot(y_periodic,'r','linewidth',2);
xlabel('Time (TR)');
title(sprintf('Responses Evoked by\nPeriodic Stimulus Onset\nVariance=%1.2f',var(y_periodic)))
subplot(122)
stem(D_random,'k');
hold on;
plot(y_random,'r','linewidth',2);
xlabel('Time (TR)');
title(sprintf('Responses Evoked by\nRandom Stimulus Onset\nVariance=%1.2f',var(y_random)))
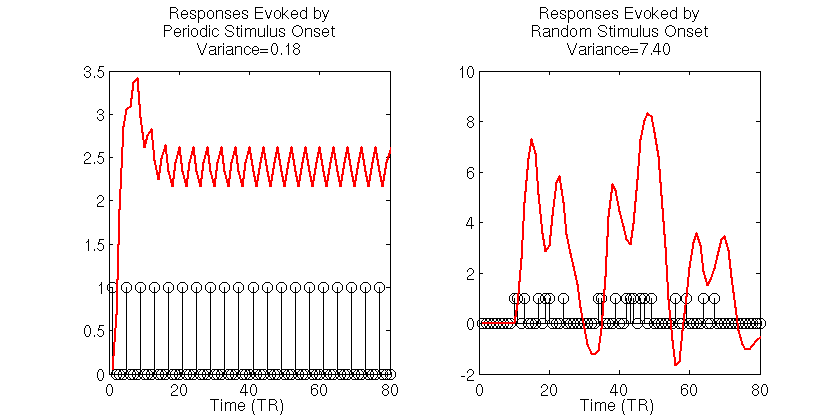
The black stick functions in the simulation output indicate the stimulus onsets and each red function is the simulated noiseless BOLD signal to those stimuli. The first thing to notice is the dramatically different variances of the BOLD signals evoked for the two stimulus presentation schedules. For the periodic stimuli, the BOLD signal quickly saturates, then oscillates around an effective baseline activation. The estimated variance of the periodic-based signal is 0.18. In contrast, the signal evoked by the random stimulus presentation schedule varies wildly, reaching a maximum amplitude that is roughly 2.5 times as large the maximum amplitude of the signal evoked by periodic stimuli. The estimated variance of the signal evoked by the random stimuli is 7.4, roughly 40 times the variance of the signal evoked by the periodic stimulus.
So which stimulus schedule allows us to better estimate the HRF and, more importantly, the amplitude of the HRF, as it is the amplitude that is the common proxy for voxel selectivity/activation? Below we repeat the above experiment 50 times. However, instead of simulating noiseless BOLD responses, we introduce 50 distinct, uncorrelated noise conditions, and from the simulated noisy responses, we estimate the HRF using an FIR basis set for each repeated trial. We then compare the estimated HRFs across the 50 trials for the periodic and random stimulus presentation schedules. Note that for each trial, the noise is exactly the same for the two stimulus presentation schedules. Further, we simulate a selectivity/tuning gain of 3 times the maximum HRF amplitude and assume that the HRF to be estimated is 16 TRs/seconds in length. The simulation and output are below:
%% SIMULATE MULTIPLE TRIALS OF EACH EXPERIMENT
%% AND ESTIMATE THE HRF FOR EACH
%% (ASSUME THE VARIABLES DEFINED ABOVE ARE IN WORKSPACE)
% CREATE AN FIR DESIGN MATRIX
% FOR EACH EXPERIMENT
hrfLen = 16; % WE ASSUME TO-BE-ESTIMATED HRF IS 16 TRS LONG
% CREATE FIR DESIGN MATRIX FOR THE PERIODIC STIMULI
X_FIR_periodic = zeros(nTRs,hrfLen);
onsets = find(D_periodic);
idxCols = 1:hrfLen;
for jO = 1:numel(onsets)
idxRows = onsets(jO):onsets(jO)+hrfLen-1;
for kR = 1:numel(idxRows);
X_FIR_periodic(idxRows(kR),idxCols(kR)) = 1;
end
end
X_FIR_periodic = X_FIR_periodic(1:nTRs,:);
% CREATE FIR DESIGN MATRIX FOR THE RANDOM STIMULI
X_FIR_random = zeros(nTRs,hrfLen);
onsets = find(D_random);
idxCols = 1:hrfLen;
for jO = 1:numel(onsets)
idxRows = onsets(jO):onsets(jO)+hrfLen-1;
for kR = 1:numel(idxRows);
X_FIR_random(idxRows(kR),idxCols(kR)) = 1;
end
end
X_FIR_random = X_FIR_random(1:nTRs,:);
% SIMULATE AND ESTIMATE HRF WEIGHTS VIA OLS
nTrials = 50;
% CREATE NOISE TO ADD TO SIGNALS
% NOTE: SAME NOISE CONDITIONS FOR BOTH EXPERIMENTS
noiseSTD = beta*2;
noise = bsxfun(@times,randn(nTrials,numel(X_periodic)),noiseSTD);
%% ESTIMATE HRF FROM PERIODIC STIMULUS TRIALS
beta_periodic = zeros(nTrials,hrfLen);
for iT = 1:nTrials
y = y_periodic + noise(iT,:);
beta_periodic(iT,:) = X_FIR_periodic\y';
end
% CALCULATE MEAN AND STANDARD ERROR OF HRF ESTIMATES
beta_periodic_mean = mean(beta_periodic);
beta_periodic_se = std(beta_periodic)/sqrt(nTrials);
%% ESTIMATE HRF FROM RANDOM STIMULUS TRIALS
beta_random = zeros(nTrials,hrfLen);
for iT = 1:nTrials
y = y_random + noise(iT,:);
beta_random(iT,:) = X_FIR_random\y';
end
% CALCULATE MEAN AND STANDARD ERROR OF HRF ESTIMATES
beta_random_mean = mean(beta_random);
beta_random_se = std(beta_random)/sqrt(nTrials);
% DISPLAY HRF ESTIMATES
figure
% ...FOR THE PERIODIC STIMULI
subplot(121);
hold on;
h0 = plot(h*beta,'k')
h1 = plot(beta_periodic_mean,'linewidth',2);
h2 = plot(beta_periodic_mean+beta_periodic_se,'r','linewidth',2);
plot(beta_periodic_mean-beta_periodic_se,'r','linewidth',2);
xlabel('Time (TR)')
legend([h0, h1,h2],'Actual HRF','Average \beta_{periodic}','Standard Error')
title('Periodic HRF Estimate')
% ...FOR THE RANDOMLY-PRESENTED STIMULI
subplot(122);
hold on;
h0 = plot(h*beta,'k');
h1 = plot(beta_random_mean,'linewidth',2);
h2 = plot(beta_random_mean+beta_random_se,'r','linewidth',2);
plot(beta_random_mean-beta_random_se,'r','linewidth',2);
xlabel('Time (TR)')
legend([h0,h1,h2],'Actual HRF','Average \beta_{random}','Standard Error')
title('Random HRF Estimate')
In the simulation outputs, the average HRF for the random stimulus presentation (right) closely follows the actual HRF tuning. Also, there is little variability of the HRF estimates, as is indicated by the small standard error estimates for each time points. As well, the selectivity/gain term is accurately recovered, giving a mean HRF with nearly the same amplitude as the underlying model. In contrast, the HRF estimated from the periodic-based experiment is much more variable, as indicated by the large standard error estimates. Such variability in the estimates of the HRF reduce our confidence in the estimate for any single trial. Additionally, the scale of the mean HRF estimate is off by nearly 30% of the actual value.
From these results, it is obvious that the random stimulus presentation rate gives rise to more accurate, and less variable estimates of the HRF function. What may not be so obvious is why this is the case, as there were the same number of stimuli and the same number of signal measurements in each experiment. To get a better understanding of why this is occurring, let’s refer back to the variances of the evoked noiseless signals. These are the signals that are underlying the noisy signals used to estimate the HRF. When noise is added it impedes the detection of the underlying trends that are useful for estimating the HRF. Thus it is important that the variance of the underlying signal is large compared to the noise so that the signal can be detected.
For the periodic stimulus presentation schedule, we saw that the variation in the BOLD signal was much smaller than the variation in the BOLD signals evoked during the randomly-presented stimuli. Thus the signal evoked by random stimulus schedule provide a better characterization of the underlying signal in the presence of the same amount of noise, and thus provide more information to estimate the HRF. With this in mind we can think of maximizing the efficiency of the an experiment design as maximizing the variance of the BOLD signals evoked by the experiment.
An Alternative Perspective: The Frequency Power Spectrum
Another helpful interpretation is based on a signal processing perspective. If we assume that neural activity is directly correspondent with the onset of a stimulus event, then we can interpret the train of stimulus onsets as a direct signal of the evoked neural activity. Furthermore, we can interpret the HRF as a low-pass-filter that acts to “smooth” the available neural signal in time. Each of these signals–the neural/stimulus signal and the HRF filtering signal–has with it an associated power spectrum. The power spectrum for a signal captures the amount of power per unit time that the signal has as a particular frequency . The power spectrum for a discrete signal can be calculated from the discrete Fourier transform (DFT) of the signal
as follows
Below, we use Matlab’s function to calculate the DFT and the associated power spectrum for each of the stimulus/neural signals, as well as the HRF.
%% POWER SPECTRUM ANALYSES
%% (ASSUME THE VARIABLES DEFINED ABOVE ARE IN WORKSPACE)
% MAKE SURE WE PAD SUFFICIENTLY
% FOR CIRCULAR CONVOLUTION
N = 2^nextpow2(nTRs + numel(h)-1);
nUnique = ceil(1+N/2); % TAKE ONLY POSITIVE SPECTRA
% CALCULATE POWER SPECTRUM FOR PERIODIC STIMULI EXPERIMENT
ft_D_periodic = fft(D_periodic,N)/N; % DFT
P_D_periodic = abs(ft_D_periodic).^2; % POWER
P_D_periodic = 2*P_D_periodic(2:nUnique-1); % REMOVE ZEROTH & NYQUIST
% CALCULATE POWER SPECTRUM FOR RANDOM STIMULI EXPERIMENT
ft_D_random = fft(D_random,N)/N; % DFT
P_D_random = abs(ft_D_random).^2; % POWER
P_D_random = 2*P_D_random(2:nUnique-1); % REMOVE ZEROTH & NYQUIST
% CALCULATE POWER SPECTRUM OF HRF
ft_h = fft(h,N)/N; % DFT
P_h = abs(ft_h).^2; % POWER
P_h = 2*P_h(2:nUnique-1); % REMOVE ZEROTH & NYQUIST
% CREATE A FREQUENCY SPACE FOR PLOTTING
F = 1/N*[1:N/2-1];
% DISPLAY STIMULI POWER SPECTRA
figure
subplot(131)
hhd = plot(F,P_D_periodic,'b','linewidth',2);
axis square; hold on;
hhr = plot(F,P_D_random,'g','linewidth',2);
xlim([0 .3]); xlabel('Frequency (Hz)');
set(gca,'Ytick',[]); ylabel('Magnitude');
legend([hhd,hhr],'Periodic','Random')
title('Stimulus Power, P_{stim}')
% DISPLAY HRF POWER SPECTRUM
subplot(132)
plot(F,P_h,'r','linewidth',2);
axis square
xlim([0 .3]); xlabel('Frequency (Hz)');
set(gca,'Ytick',[]); ylabel('Magnitude');
title('HRF Power, P_{HRF}')
% DISPLAY EVOKED SIGNAL POWER SPECTRA
subplot(133)
hhd = plot(F,P_D_periodic.*P_h,'b','linewidth',2);
hold on;
hhr = plot(F,P_D_random.*P_h,'g','linewidth',2);
axis square
xlim([0 .3]); xlabel('Frequency (Hz)');
set(gca,'Ytick',[]); ylabel('Magnitude');
legend([hhd,hhr],'Periodic','Random')
title('Signal Power, P_{stim}.*P_{HRF}')
On the left of the output we see the power spectra for the stimulus signals. The blue line corresponds to the spectrum for the periodic stimuli, and the green line the spectrum for the randomly-presented stimuli. The large peak in the blue spectrum corresponds to the majority of the stimulus power at 0.25 Hz for the periodic stimuli, as this the fundamental frequency of the periodic stimulus presentation (i.e. every 4 seconds). However, there is little power at any other stimulus frequencies. In contrast the green spectrum indicates that the random stimulus presentation has power at multiple frequencies.
If we interpret the HRF as a filter, then we can think of the HRF power spectrum as modulating the power spectrum of the neural signals to produce the power of the evoked BOLD signals. The power spectrum for the HRF is plotted in red in the center plot. Notice how a majority of the power for the HRF is at frequencies less than 0.1 Hz, and there is very little power at frequencies above 0.2 Hz. If the neural signal power is modulated by the HRF signal power, we see that there is little resultant power in the BOLD signals evoked by periodic stimulus presentation (blue spectrum in the right plot). In contrast, because the power for the neural signals evoked by random stimuli are spread across the frequency domain, there are a number of frequencies that overlap with those frequencies for which the HRF also has power. Thus after modulating neural/stimulus power with the HRF power, the spectrum of the BOLD signals evoked by the randomly-presented stimuli have much more power across the relevant frequency spectrum than those evoked by the periodic stimuli. This is indicated by the larger area under the green curve in the right plot.
Using the signal processing perspective allows us to directly gain perspective on the limitations of a particular experiment design which are rooted in the frequency spectrum of the HRF. Therefore, another way we can think of maximizing the efficiency of an experimental design is maximizing the amount of power in the resulting evoked BOLD responses.
Yet Another Perspective Based in Statistics: Efficiency Metric
Taking a statistics-based approach leads to a formal definition of efficiency, and further, a nice metric for testing the efficiency of an experimental design. Recall that when determining the shape of the HRF, a common approach is to use the GLM model
Here is the evoked BOLD signal and
is a design matrix that links a set of linear model parameters
to those responses. The variable
is a noise term that is unexplained by the model. Using an FIR basis formulation of the model, the weights in
represent the HRF to a stimulus condition.
Because fMRI data are a continuous time series, the underlying noise is generally correlated in time. We can model this noise as a Gaussian process with zero mean and a constant multivariate covariance
. Note that this is analogous to the Generalized Least Squares (GLS) formulation of the GLM. In general, the values that comprise
are unknown and have to be estimated from the fMRI data themselves.
For a known or estimated noise covariance, the Maximum Likelihood Estimator (MLE) for the model parameters (derivation not shown) is:
Because the ML estimator of the HRF is a linear combination of the design matrix and a set of corresponding responses, which are both random variables (
can represent any possible experiment design, and
is by definition random), the estimator is itself a random variable. It thus follows that the estimate for the HRF also has a variance. (We demonstrated how
is a random variable in the 50 simulations above, where for each simulation X was held fixed, but due to the added noise
was a random variable. For each noise condition, the estimate for
took on different values.) We saw above how an HRF estimator with a large variance is undesirable, as it reduces our confidence in the estimates of the HRF shape and scale. Therefore we would like to determine an estimator that has a minimum overall variance.
A formal metric for efficiency of a least-squares estimator is directly related to the variance of the estimator. The efficiency is defined to be the inverse of the sum of the estimator variances. An estimator that has a large sum of variances will have a low efficiency, and vice versa. But how do we obtain the values of the variances for the estimator? The variances can be recovered from the diagonal elements of the estimator covariance matrix , giving the following definition for the efficiency,
In earlier post we found that the covariance matrix for the GLS estimator (i.e. the formulation above) with a given noise covariance
is:
.
Thus the efficiency for the HRF estimator is
Here we see that the efficiency depends only on the known noise covariance (or an estimate of it), and the design matrix used in the model, but not the shape of the HRF. In general the noise covariance is out of the experimenter’s control (but see the take-homes below ), and must be dealt with post hoc. However, because the design matrix is directly related to the experimental design, the above expression gives a direct way to test the efficiency of experimental designs before they are ever used!
In the simulations above, the noise processes are drawn from an independent multivariate Gaussian distribution, therefore the noise covariance is equal to the identity (i.e. uncorrelated). We also estimated the HRF using the FIR basis set, thus our model design matrix was . This gives the estimate the efficiency for the simulation experiments:
Below we calculate the efficiency for the FIR estimates under the simulated experiments with periodic and random stimulus presentation designs.
%% ESTIMATE DESIGN EFFICIENCY
%% (ASSUME THE VARIABLES DEFINED ABOVE ARE IN WORKSPACE)
% CALCULATE EFFICIENCY OF PERIODIC EXPERIMENT
E_periodic = 1/trace(pinv(X_FIR_periodic'*X_FIR_periodic));
% CALCULATE EFFICIENCY OF RANDOM EXPERIMENT
E_random = 1/trace(pinv(X_FIR_random'*X_FIR_random));
% DISPLAY EFFICIENCY ESTIMATES
figure
bar([E_periodic,E_random]);
set(gca,'XTick',[1,2],'XTickLabel',{'E_periodic','E_random'});
title('Efficiency of Experimental Designs');
colormap hot;
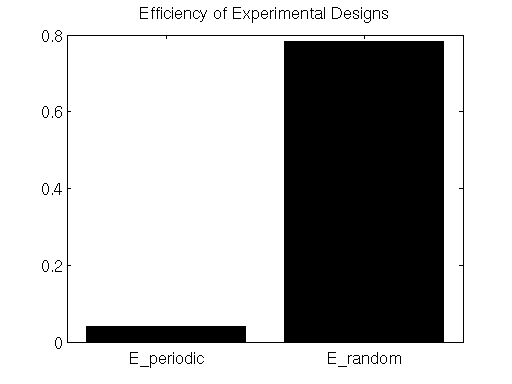
Here we see that the efficiency metric does indeed indicate that the randomly-presented stimulus paradigm is far more efficient than the periodically-presented paradigm.
Wrapping Up
In this post we addressed the efficiency of an fMRI experiment design. A few take-homes from the discussion are:
- Randomize stimulus onset times. These onset times should take into account the low-pass characteristics (i.e. the power spectrum) of the HRF.
- Try to model selectivity to events that occur close in time. The reason for this is that noise covariances in fMRI are highly non-stationary. There are many sources of low-frequency physiological noise such as breathing, pulse, blood pressure, etc, all of which dramatically effect the noise in the fMRI timecourses. Thus any estimate of noise covariances from data recorded far apart in time will likely be erroneous.
- Check an experimental design against other candidate designs using the Efficiency metric.
Above there is mention of the effects of low-frequency physiological noise. Until now, our simulations have assumed that all noise is independent in time, greatly simplifying the picture of estimating HRFs and corresponding selectivity. However, in a later post we’ll address how to deal with more realistic time courses that are heavily influenced by sources of physiological noise. Additionally, we’ll tackle how to go about estimating the noise covariance from more realistic fMRI time series.