Monthly Archives: March 2013
The Statistical Whitening Transform
In a number of modeling scenarios, it is beneficial to transform the to-be-modeled data such that it has an identity covariance matrix, a procedure known as Statistical Whitening. When data have an identity covariance, all dimensions are statistically independent, and the variance of the data along each of the dimensions is equal to one. (To get a better idea of what an identity covariance entails, see the following post.)
Enforcing statistical independence is useful for a number of reasons. For example, in probabilistic models of data that exist in multiple dimensions, the joint distribution–which may be very complex and difficult to characterize–can factorize into a product of many simpler distributions when the dimensions are statistically independent. Forcing all dimensions to have unit variance is also useful. For instance, scaling all variables to have the same variance treats each dimension with equal importance.
In the remainder of this post we derive how to transform data such that it has an identity covariance matrix, give some examples of applying such a transformation to real data, and address some interpretations of statistical whitening in the scope of theoretical neuroscience.
Decorrelation: Transforming Data to Have a Diagonal Covariance Matrix
Let’s say we have some data matrix composed of
dimensions and
observations (
has size
). Let’s also assume that the rows of
have been centered (the mean has been subracted across all observations) . The covariance
of each of the dimensions with respect to the other is
(1)
Where the covariance can be estimated from the data matrix as follows:
(2)
The covariance matrix , by definition (Equation 2) is symmetric and positive semi-definite (if you don’t know what that means, don’t worry it’s not terribly important for this discussion). Thus we can write the matrix as the product of two simpler matrices
and
, using a procedure known as Eigenvalue Decomposition:
(3)
The matrix is an
-sized matrix, where each column is an eigenvector of
, and
is a diagonal matrix whose diagonal elements
are eigenvalues that correspond to the eigenvectors of the
-th column of
. For more details on eigenvectors and eigenvalues see the following. From Equation (3), and using a little algebra, we can transform
into the diagonal matrix
(4)
Now, imagine the goal is to transform the data matrix into a new data matrix
(5)
whose dimensions are uncorrelated (i.e. has a diagonal covariance
). Thus we want to determine the transformation
that makes:
(6)
Here we derive the expression for using Equations (2), (4), (5), and (6):
(a la Equations (5) and (6))
(via Equation (2))
(via Equation (4))
now, because (see following link for details)
and thus
(7)
This means that we can transform into an uncorrelated (i.e. orthogonal) set of variables by premultiplying data matrix
with the transpose of the the eigenvectors of data covariance matrix
.
Whitening: Transforming data to have an Identity Covariance matrix
Ok, so now we have a way of transforming our data so that the dimensions are uncorrelated. However, this only gives us a diagonal covariance matrix, not an Identity covariance matrix. In order to obtain an Identity covariance, we also need to scale each dimension so that its variance is equal to one. How can we determine this transformation? We know how to transform our data so that the covariance is equal to . If we can determine the transformation that leaves
, then we can apply this transformation to our decorrelated covariance to give us the desired whitening transform. We can determine this from the somewhat trivial notion that
(8)
and further that
(9)
Now, using Equation (4) along with Equation (8), we can see that
(10)
Now say that we define a variable , where
is the desired whitening transform, that leaves the covariance of
equal to the identity matrix. Using essentially the same set of derivation steps as above to solve for
, but starting from Equation (9) we find that
(11)
(12)
Thus, the whitening transform is simply the decorrelation transform, but scaled by the inverse of the square root of the (here the inverse and square root can be performed element-wise because
is a diagonal matrix).
Interpretation of the Whitening Transform
So what does the whitening transformation actually do to the data (below, blue points)? We investigate this transformation below: The first operation decorrelates the data by premultiplying the data with the eigenvector matrix , calculated from the data covariance. This decorrelation can be thought of as a rotation that reorients the data so that the principal axes of the data are aligned with the axes along which the data has the largest (orthogonal) variance. This rotation is essentially the same procedure as the oft-used Principal Components Analysis (PCA), and is shown in the middle row.
The second operation, scaling by can be thought of squeezing the data–if the variance along a dimension is larger than one–or stretching the data–if the variance along a dimension is less than one. The stretching and squeezing forms the data into a sphere about the origin (which is why whitening is also referred to as “sphering”). This scaling operation is depicted in the bottom row in the plot above.
The MATLAB to make make the plot above is here:
% INITIALIZE SOME CONSTANTS
mu = [0 0];
S = [1 .9; .9 3];
% SAMPLE SOME DATAPOINTS
nSamples = 1000;
samples = mvnrnd(mu,S,nSamples)';
% WHITEN THE DATA POINTS...
[E,D] = eig(S);
% ROTATE THE DATA
samplesRotated = E*samples;
% TAKE D^(-1/2)
D = diag(diag(D).^-.5);
% SCALE DATA BY D
samplesRotatedScaled = D*samplesRotated;
% DISPLAY
figure;
subplot(311);
plot(samples(1,:),samples(2,:),'b.')
axis square, grid
xlim([-5 5]);ylim([-5 5]);
title('Original Data');
subplot(312);
plot(samplesRotated(1,:),samplesRotated(2,:),'r.'),
axis square, grid
xlim([-5 5]);ylim([-5 5]);
title('Decorrelate: Rotate by V');
subplot(313);
plot(samplesRotatedScaled(1,:),samplesRotatedScaled(2,:),'ko')
axis square, grid
xlim([-5 5]);ylim([-5 5]);
title('Whiten: scale by D^{-1/2}');
The transformation in Equation (11) and implemented above whitens the data but leaves the data aligned with principle axes of the original data. In order to observe the data in the original space, it is often customary “un-rotate” the data back into it’s original space. This is done by just multiplying the whitening transform by the inverse of the rotation operation defined by the eigenvector matrix. This gives the whitening transform:
(13)
Let’s take a look an example of using statistical whitening for a more complex problem: whitening patches of images sampled from natural scenes.
Example: Whitening Natural Scene Image Patches
Modeling the local spatial structure of pixels in natural scene images is important in many fields including computer vision and computational neuroscience. An interesting model of natural scenes is one that can account for interesting, high-order statistical dependencies between pixels. However, because natural scenes are generally composed of continuous objects or surfaces, a vast majority of the spatial correlations in natural image data can be explained by local pairwise dependencies. For example, observe the image below.
% LOAD AND DISPLAY A NATURAL IMAGE
im = double(imread('cameraman.tif'));
figure
imagesc(im); colormap gray; axis image; axis off;
title('Base Image')
Given one of the gray pixels in the upper portion of the image, it is very likely that all pixels within the local neighborhood will also be gray. Thus there is a large amount of correlation between pixels in local regions of natural scenes. Statistical models of local structure applied to natural scenes will be dominated by these pairwise correlations, unless they are removed by preprocessing. Whitening provides such a preprocessing procedure.
Below we create and display a dataset of local image patches of size extracted at random from the image above. Each patch is rastered out into a column vector of size
. Each of these patches can be thought of as samples of the local structure of this natural scene. Below we use the whitening transformation to remove pairwise correlations between pixels in each patch and scale the variance of each pixel to be one.
On the left is the dataset of extracted image patches, along with the corresponding covariance matrix for the image patches on the right. The large local correlation within the neighborhood of each pixel is indicated by the large bright diagonal regions throughout the covariance matrix.
The MATLAB code to extract and display the patches shown above is here:
% CREATE PATCHES DATASET FROM NATURAL IMAGE
rng(12345)
imSize = 256;
nPatches = 400; % (MAKE SURE SQUARE)
patchSize = 16;
patches = zeros(patchSize*patchSize,nPatches);
patchIm = zeros(sqrt(nPatches)*patchSize);
% PAD IMAGE FOR EDGE EFFECTS
im = padarray(im,[patchSize,patchSize],'symmetric');
% EXTRACT PATCHES...
for iP = 1:nPatches
pix = ceil(rand(2,1)*imSize);
rows = pix(1):pix(1)+patchSize-1;
cols = pix(2):pix(2)+patchSize-1;
tmp = im(rows,cols);
patches(:,iP) = reshape(tmp,patchSize*patchSize,1);
rowIdx = (ceil(iP/sqrt(nPatches)) - 1)*patchSize + ...
1:ceil(iP/sqrt(nPatches))*patchSize;
colIdx = (mod(iP-1,sqrt(nPatches)))*patchSize+1:patchSize* ...
((mod(iP-1,sqrt(nPatches)))+1);
patchIm(rowIdx,colIdx) = tmp;
end
% CENTER IMAGE PATCHES
patchesCentered = bsxfun(@minus,patches,mean(patches,2));
% CALCULATE COVARIANCE MATRIX
S = patchesCentered*patchesCentered'/nPatches;
% DISPLAY PATCHES
figure;
subplot(121);
imagesc(patchIm);
axis image; axis off; colormap gray;
title('Extracted Patches')
% DISPLAY COVARIANCE
subplot(122);
imagesc(S);
axis image; axis off; colormap gray;
title('Extracted Patches Covariance')
Below we implement the whitening transformation described above to the extracted image patches and display the whitened patches that result.
 On the left, we see that the whitening procedure zeros out all areas in the extracted patches that have the same value (zero is indicated by gray). The whitening procedure also boosts the areas of high-contrast (i.e. edges). The right plots the covariance matrix for the whitened patches. The covarance matrix is diagonal, indicating that pixels are now independent. In addition, all diagonal entries have the same value, indicating the that all pixels now have the same variance (i.e. 1). The MATLAB code used to whiten the image patches and create the display above is here:
On the left, we see that the whitening procedure zeros out all areas in the extracted patches that have the same value (zero is indicated by gray). The whitening procedure also boosts the areas of high-contrast (i.e. edges). The right plots the covariance matrix for the whitened patches. The covarance matrix is diagonal, indicating that pixels are now independent. In addition, all diagonal entries have the same value, indicating the that all pixels now have the same variance (i.e. 1). The MATLAB code used to whiten the image patches and create the display above is here:
%% MAIN WHITENING
% DETERMINE EIGENECTORS & EIGENVALUES
% OF COVARIANCE MATRIX
[E,D] = eig(S);
% CALCULATE D^(-1/2)
d = diag(D);
d = real(d.^-.5);
D = diag(d);
% CALCULATE WHITENING TRANSFORM
W = E*D*E';
% WHITEN THE PATCHES
patchesWhitened = W*patchesCentered;
% DISPLAY THE WHITENED PATCHES
wPatchIm = zeros(size(patchIm));
for iP = 1:nPatches
rowIdx = (ceil(iP/sqrt(nPatches)) - 1)*patchSize + 1:ceil(iP/sqrt(nPatches))*patchSize;
colIdx = (mod(iP-1,sqrt(nPatches)))*patchSize+1:patchSize* ...
((mod(iP-1,sqrt(nPatches)))+1);
wPatchIm(rowIdx,colIdx) = reshape(patchesWhitened(:,iP),...
[patchSize,patchSize]);
end
figure
subplot(121);
imagesc(wPatchIm);
axis image; axis off; colormap gray; caxis([-5 5]);
title('Whitened Patches')
subplot(122);
imagesc(cov(patchesWhitened'));
axis image; axis off; colormap gray; %colorbar
title('Whitened Patches Covariance');
Investigating the Whitening Matrix: implications for theoretical neuroscience
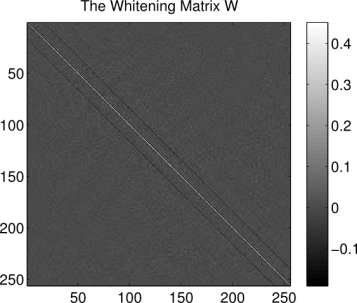
So what does the whitening matrix look like, and what does it do? Below is the whitening matrix calculated for the image patches dataset:
% DISPLAY THE WHITENING MATRIX
figure; imagesc(W);
axis image; colormap gray; colorbar
title('The Whitening Matrix W')
Each column of is the operation that scales the variance of the corresponding pixel to be equal to one and forces that pixel independent of the others in the
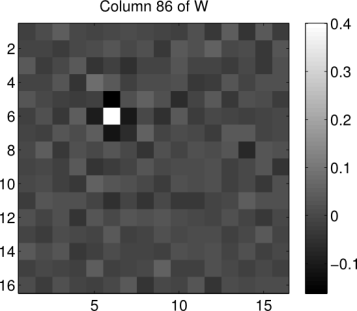
patch. So what exactly does such an operation look like? We can get an idea by reshaping a column of W back into the shape of the image patches. Below we show what the 86th column of W looks like when reshaped in such a way (the index 86 has no particular significance, it was chosen at random):
% DISPLAY A COLUMN OF THE WHITENING MATRIX
figure; imagesc(reshape(W(:,86),16,16)),
colormap gray,
axis image, colorbar
title('Column 86 of W')
We see that the operation is essentially an impulse centered on the 86th pixel in the image (counting pixels starting in the upper left corner, proceeding down columns). This impulse is surrounded by inhibitory weights. If we were to look at the remaining columns of , we would find that that the same center-surround operation is being replicated at every pixel location in each image patch. Essentially, the whitening transformation is performing a convolution of each image patch with a center-surround filter whose properties are estimated from the patches dataset. Similar techniques are common in computer vision edge-detection algorithms.
Implications for theoretical neuroscience
A theoretical function of the primate retina is data compression: a large number of photoreceptors pass data from the retina into a physiological bottleneck, the optic nerve, which has far fewer fibers than retinal photoreceptors. Thus removing redundant information is an important task that the retina must perform. When observing the whitened image patches above, we see that redundant information is nullified; pixels that have similar local values to one another are zeroed out. Thus, statistical whitening is a viable form of data compression
It turns out that there is a large class of ganglion cells in the retina whose spatial receptive fields exhibit…that’s right center-surround activation-inhibition like the operation of the whitening matrix shown above! Thus it appears that the primate visual system may be performing data compression at the retina by means of a similar operation to statistical whitening. Above, we derived the center-surround whitening operation based on data sampled from a natural scene. Thus it is seems reasonable that the primate visual system may have evolved a similar data-compression mechanism through experience with natural scenes, either through evolution, or development.
Covariance Matrices and Data Distributions
Correlation between variables in a -dimensional dataset are often summarized by a
covariance matrix. To get a better understanding of how correlation matrices characterize correlations between data points, we plot data points drawn from 3 different 2-dimensional Gaussian distributions, each of which is defined by a different covariance matrix.
The left plots below display the covariance matrix for each Gaussian distribution. The values along the diagonal represent the variance of the data along each dimension, and the off-diagonal values represent the covariances between the dimensions. Thus the
-th entry of each matrix represents the correlation between the
-th and
-th dimensions. The right plots show data drawn from the corresponding 2D Gaussian.
The top row plot display a covariance matrix equal to the identity matrix, and the points drawn from the corresponding Gaussian distribution. The diagonal values are 1, indicating the data have variance of 1 along both of the dimensions. Additionally, the off-diagonal elements are zero, meaning that the two dimensions are uncorrelated. We can see this in the data drawn from the distribution as well. The data are distributed in a sphere about origin. For such a distribution of points, it is difficult (impossible) to draw any single regression line that can predict the second dimension from the first, and vice versa. Thus an identity covariance matrix is equivalent to having independent dimensions, each of which has unit (i.e. 1) variance. Such a dataset is often called “white” (this naming convention comes from the notion that white noise signals–which can be sampled from independent Gaussian distributions–have equal power at all frequencies in the Fourier domain).
The middle row plots the points that result from a diagonal, but not identity covariance matrix. The off-diagonal elements are still zero, indicating that the dimensions are uncorrelated. However, the variances along each dimension are not equal to one, and are not equal. This is demonstrated by the elongated distribution in red. The elongation is along the second dimension, as indicated by the larger value in the bottom-right (point ) of the covariance matrix.
The bottom row plots points that result from a non-diagonal covariance matrix. Here the off-diagonal elements of covariance matrix have non-zero values, indicating a correlation between the dimensions. This correlation is reflected in the distribution of drawn datapoints (in blue). We can see that the primary axis along which the points are distributed is not along either of the dimensions, but a linear combination of the dimensions.
The MATLAB code to create the above plots is here
% INITIALIZE SOME CONSTANTS
mu = [0 0]; % ZERO MEAN
S = [1 .9; .9 3]; % NON-DIAGONAL COV.
SDiag = [1 0; 0 3]; % DIAGONAL COV.
SId = eye(2); % IDENTITY COV.
% SAMPLE SOME DATAPOINTS
nSamples = 1000;
samples = mvnrnd(mu,S,nSamples)';
samplesId = mvnrnd(mu,SId,nSamples)';
samplesDiag = mvnrnd(mu,SDiag,nSamples)';
% DISPLAY
subplot(321);
imagesc(SId); axis image,
caxis([0 1]), colormap hot, colorbar
title('Identity Covariance')
subplot(322)
plot(samplesId(1,:),samplesId(2,:),'ko'); axis square
xlim([-5 5]), ylim([-5 5])
grid
title('White Data')
subplot(323);
imagesc(SDiag); axis image,
caxis([0 3]), colormap hot, colorbar
title('Diagonal Covariance')
subplot(324)
plot(samplesDiag(1,:),samplesDiag(2,:),'r.'); axis square
xlim([-5 5]), ylim([-5 5])
grid
title('Uncorrelated Data')
subplot(325);
imagesc(S); axis image,
caxis([0 3]), colormap hot, colorbar
title('Non-diagonal Covariance')
subplot(326)
plot(samples(1,:),samples(2,:),'b.'); axis square
xlim([-5 5]), ylim([-5 5])
grid
title('Correlated Data')